and
installing it.
There is a
very good set of video tutorials with source code here that take
about 2-3 hours to complete:
https://shiny.rstudio.com/tutorial/ that will give you a very good
grounding in the basics.
There is also Swirl for learning R - http://swirlstats.com/ which
is also a very nice way to learn the basics of R.
We will be using R for all of the projects so its worth taking the
time to learn it now.
Some useful links
Start RStudio and you should see something similar to a
modern IDE with an area for viewing and editing code, a
terminal, an area for seeing the current state of the R
environment, and a multi-purpose area.
A lot of the power of R comes from the variety of packages
that can be installed into it
To see what packages are currently installed, in the terminal
at the > prompt, type installed.packages()[,1:2]
To see which packages are out of date type old.packages()
To update all old packages type update.packages(ask =
FALSE)
Some useful libraries to install with install.packages()
include:
- shiny
- shinydashboard
- ggplot2
- lubridate
- DT
- jpeg
- leaflet
e.g.
install.packages("shiny")
I will be going through a demonstration of using these tools to
visualize some local temperature data in class. For over 15
years we have been collecting temperature data in the various
rooms of the lab, trying to understand why the temperature used
to change so dramatically at times, trying to keep the people
and the machines here happy.
The relevant files are located
here
What this code produces is interactive, hosted on the shiny
site,
here
and a snapshot below:
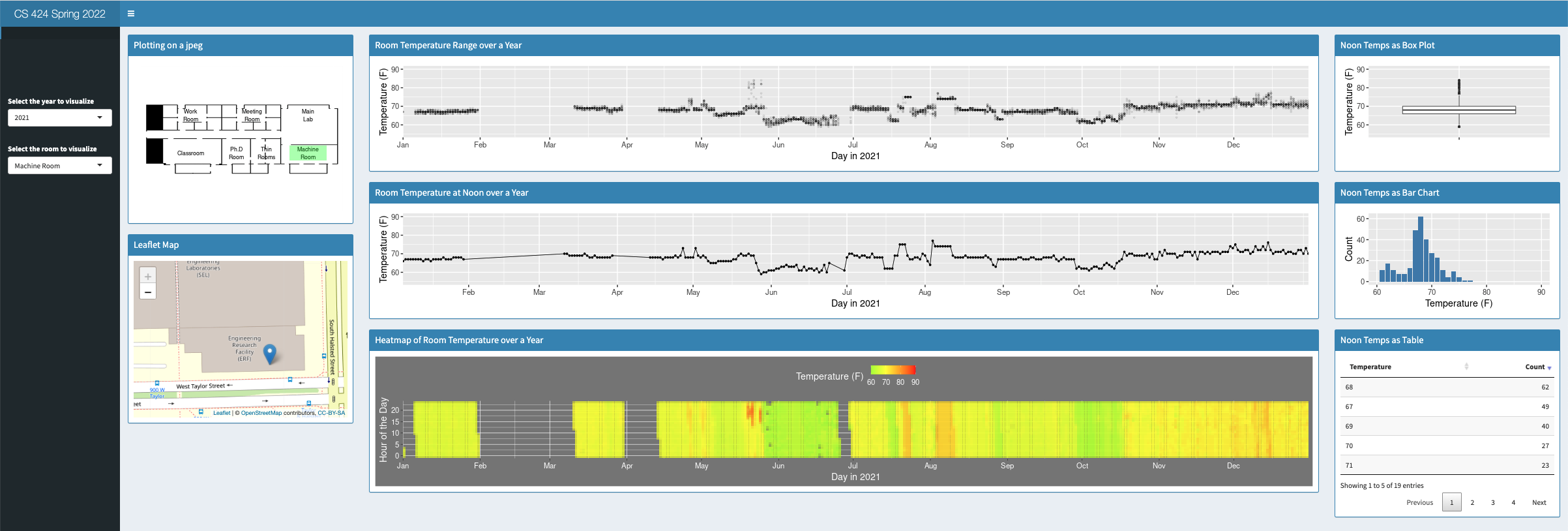
The visualization was designed to run as a compromise between an
HD display and our wide classroom wall and in either case be
touch-screen compatible, which is also why the version you
download has the menu at the upper left slightly lowered so
people could reach it to interact with it at the wall. The
projects will involve developing and demonstrating
visualizations for this classroom wall.
 Lets play a bit with the evl temperature
data using R and R studio. As you go through this tutorial
take a screen snapshot including each of the visualizations.
You will be turning these in through gradescope.
Lets play a bit with the evl temperature
data using R and R studio. As you go through this tutorial
take a screen snapshot including each of the visualizations.
You will be turning these in through gradescope.
one nice way to do this is to copy and paste the following
relevant commands below into the RStudio console (in the lower
left panel of R studio) one by one to see their affect on the
current environment.
download a copy of the evlWeatherForR.zip file from the
'relevant files' link above and unzip it.
set the working directory to the evlWeatherForR directory
setwd("dir-you-want-to-go-to
")
test if thats correct with
getwd()
to get a listing of the directory use
dir()
read in one file
evl2006 <- read.table(file = "history_2006.tsv", sep =
"\t", header = TRUE) ... be careful to avoid smart
quotes. You should now see evl2006 in the Global Environment at
the upper right
take a look at it
evl2006
and then some commands to get an overall picture of the data:
str(evl2006)
summary(evl2006)
head(evl2006)
tail(evl2006)
dim(evl2006)
hmmm almost all the fields are integers including the hour and
the temperature for the 7 different rooms, but Date is a factor
- what is a factor? use the Help viewer in R-Studio. R assumes
that this is categorical data and assigns an integer value to
each unique string.
convert the dates to internal format and remove the original
dates
newDates <- as.Date(evl2006$Date, "%m/%d/%Y")
evl2006$newDate<-newDates
evl2006$Date <- NULL
we didn't need to use the
intermediary newDates, but it can be safer when you are starting
out so you can check your results before you write over things
you didn't mean to. Right now our data sets are small enough
that we shouldn't need to worry about running out of memory.
if we use str(evl2006) again, now we have newDate in date format
try doing some simple graphs and stats
plot all temps in room 4 from 2006 using the built in plotting
plot(evl2006$newDate, evl2006$S4, xlab = "Month", ylab =
"Temperature")
we can set the y axis to a fixed range for all temps in a given
room
plot(evl2006$newDate, evl2006$S4, xlab = "Month", ylab =
"Temperature", ylim=c(65, 90))
the built in plotting it nice to get quick views but it isn't
very powerful or very nice looking so now lets get the noon temp
and plot it for one / all the rooms using ggplot
if ggplot2 is not already installed then lets install it
install.packages("ggplot2")
and then lets load it in
library(ggplot2)
we want values from evl2006 where $hour is 12
noons <- subset(evl2006, Hour == 12)
we can list all the noons for a particulate room
noons$S2
note that you can see similar information in the environment
panel at the upper right
or plot them
ggplot(noons, aes(x=newDate, y=S2)) +
geom_point(color="blue") + labs(title="Room Temperature
in room ???", x="Day", y = "Degrees F") + geom_line()
we can set the min and max and add some aesthetics
ggplot(noons, aes(x=newDate, y=S2)) +
geom_point(color="blue") + labs(title="Room Temperature
in room ???", x="Day", y = "Degrees F") + geom_line() +
coord_cartesian(ylim = c(65,90))
add a smooth line through the data
ggplot(noons, aes(x=newDate, y=S2)) +
geom_point(color="blue") + labs(title="Room Temperature
in room ???", x="Day", y = "Degrees F") + geom_line() +
coord_cartesian(ylim = c(65,90)) + geom_smooth()
no points just lines and the smooth curve
ggplot(noons, aes(x=newDate, y=S2)) + labs(title="Room
Temperature in room ???", x="Day", y = "Degrees F") +
geom_line() + coord_cartesian(ylim = c(65,90)) + geom_smooth()
just the smooth curve
ggplot(noons, aes(x=newDate, y=S2)) + labs(title="Room
Temperature in room ???", x="Day", y = "Degrees F") +
coord_cartesian(ylim = c(65,90)) + geom_smooth()
we can show smooth curves for all of the rooms at noon at the
same time
ggplot(noons, aes(x=newDate)) + labs(title="Room
Temperature in room ???", x="Day", y = "Degrees F") +
coord_cartesian(ylim = c(65,90)) + geom_smooth(aes(y=S2)) +
geom_smooth(aes(y=S1)) + geom_smooth(aes(y=S3)) +
geom_smooth(aes(y=S4)) + geom_smooth(aes(y=S5)) +
geom_smooth(aes(y=S6))+ geom_smooth(aes(y=S7))
show smooth curves for all of the rooms at all hours at the same
time
ggplot(evl2006, aes(x=newDate)) + labs(title="Room
Temperature in room ???", x="Day", y = "Degrees F") +
coord_cartesian(ylim = c(65,85)) + geom_smooth(aes(y=S2)) +
geom_smooth(aes(y=S1)) + geom_smooth(aes(y=S3)) +
geom_smooth(aes(y=S4)) + geom_smooth(aes(y=S5)) +
geom_smooth(aes(y=S6))+ geom_smooth(aes(y=S7))
we can play with the style of the points and make them blue
ggplot(evl2006, aes(x=newDate, y=S4)) +
geom_point(color="blue") + labs(title="Room Temperature
in room ???", x="Day", y = "Degrees F")
we can create a bar chart for all the temps for given room
ggplot(evl2006, aes(x=factor(S4))) +
geom_bar(stat="count", width=0.7, fill="steelblue")
or just the noon temps (note that we only see temps that existed
in the data so some temps may be 'missing' on the x axis (e.g.
79)
ggplot(noons, aes(x=factor(S5))) +
geom_bar(stat="count", fill="steelblue")
we can do a better bar chart that treats the temperatures as
numbers so there wont be any missing, and we can get control
over the range of the x axis.
temperatures <- as.data.frame(table(noons[,6]))
temperatures$Var1 <-
as.numeric(as.character(temperatures$Var1))
we can get a summary of the
temperature data
summary(temperatures)
ggplot(temperatures,
aes(x=Var1, y=Freq)) + geom_bar(stat="identity",
fill="steelblue") + labs(x="Temperature (F)", y = "Count") +
xlim(60,90)
and then could create a box and whisker plot of those values to
see their distribution
ggplot(temperatures, aes(x = "", y = temperatures[,1])) +
geom_boxplot() + labs(y="Temperature (F)", x="") + ylim(55,90)
in this example we had several
temperature values for a given time in each row (called "wide
data") but sometimes you get "long data" where, in this case,
each of the temperature values would be in their own row with an
identifier saying which room it is. The reshape2 library can
covert between wide and long data. In this case we can convert
the noons data with
longNoons <-
melt(data=noons, id.vars=c("Hour", "newDate"))
Now we use ggplot2 in a slightly different way, for example we can
plot all of the noon temperatures for the 7 rooms with this one
command where 'variable' is the new variable that melt created to
hold the various room identifiers.
ggplot(longNoons) +
geom_line(aes(x=newDate, y=value, color=variable))
or break them up into 7 separate
plots
ggplot(longNoons) +
geom_line(aes(x=newDate, y=value, color=variable)) +
facet_wrap(~variable)
so we have a lot of options here. Shiny allows us to give a user
access to do these things interactively on the web using a GUI.
Some things to be careful of:
- be careful of smart quotes - they are bad
- be careful of commas, especially in the shiny code
- remember to set your working directory in R Studio
- try clearing out your R studio session regularly and running
your code to make sure your code is self-contained using
rm(list=ls())
- be careful of groupings to get your lines to connect the right
way in charts
- be careful what format your data is in - certain operations
can only be performed on certain data types
Here is
another data set to play with on Thursday in class as part of a
data scavenger hunt using Jupyter
While we used R in RStudio last
time, today we are going to use R in a Jupyter Notebook. RStudio
and Shiny are nice for creating interactive applications on the
web and the IDE is very helpful for seeing information about the
data you have loaded and the functions available to you. Jupyter
is better for showing the sequence of an investigation through a
data set.
It includes Python 3.8.X in case
you don't already have it.
Then launch Anaconda-Navigator.
Then follow the tutorial here:
https://docs.anaconda.com/anaconda/navigator/tutorials/r-lang/
through step 4 and create a new R notebook by going to Home in
Anaconda Navigator, Launching Jupyter Notebook, and then in the
upper right using the New dropdown to create a new R notebook.
Note that during the creation of the new environment choosing a
version of Python other than 3.7 might cause the install to fail
for incompatible packages.
Note that Anaconda and Jupyter
are using a separate set of R libraries from the ones in R
studio. There are ways to link the two, but for now this keeps
it simple. RStudio will likely be running R 4.1.X while Anaconda
will be running 3.6.X.
A blank notebook should load.
If instead the notebook keeps trying to load the kernel for about
30 seconds, and you see issues with IRkernel in the terminal
window, and eventually you see a connection error, then we need to
add a library to R. Go back to Anaconda and click on the R
environment that you created and 'Open Terminal'. In the terminal
window that appears type 'R' at the prompt to launch the anaconda
version of R. Then at the R prompt type
install.packages('IRkernel')
and
let it install and then type
IRkernel::installspec()
and now if you launch a new Jupyter instance and create a new R
notebook in it and the kernel should load.
(if this isn't the first time you have installed or upgraded R you
may get a message that there is a prior installation of IRkernel
that can't be removed. In that case you will likely need to go and
remove the older version manually. In R you can ask where the
packages are stored with .libPaths() and then go to that location
in the file system, delete the older version, and then repeat the
install command)
In future you may need to re-launch this terminal window to update
some the packages in R.
Try to
find interesting trends and changes in those trends. One of
the main ideas here is to get a feel for how people use
visualization interactively to look for patterns and events
and outliers in the data. In this case we will start with some
familiar concepts of utility usage - electricity, water, and
natural gas.
When you are done download your Jupyter notebook as a notebook
and as an HTML file, print the HTML file to a PDF and add this
PDF to your submission to gradescope.
a quick reference:
- + adds a cell which can contain text or code
- the scissors removes a cell
- >|Run runs the line (or you can ctrl-return)
- a insert a cell above the current one
- b insert a cell below the current one
- to add a comment in a cell you change its Cell/Cell Type
from Code to Markdown
Here is some background
information on the data:
- single family house
- natural gas for heating and cooking
- has air conditioning
Some dates that you should be able to find in the data
- replaced the furnace with a more efficient model (reduces
natural gas and electricity usage)
- started serious gardening (increases water usage)
- started serious composting in the garden (reduces water
usage)
- put in low flow shower heads and low flow toilet (reduced
water usage)
- over time replaced old light bulbs with LEDs, computers
and displays became more efficient (reduces electricity
usage)
- bought an electric car, and then another one (increases
electricity usage)
- added solar panels (reduces electricity usage)
Here is how to start playing with the data:
if you want to play with a downloaded version of the data then
set the correct path to the data (as we did above) and then
utility <- read.table(file = "utilitydata2021.tsv", sep =
"\t", header = TRUE)
or read the data from the web
with
utility <- read.table(file
= "https://www.evl.uic.edu/aej/424/utilitydata2021.tsv", sep =
"\t", header = TRUE)
sometimes there is missing data - lets check
complete.cases(utility) - all of the rows should be TRUE,
so we are good. If not, then:
utility[complete.cases(utility), ]
utility <- utility[complete.cases(utility), ]
at this point you should get a
whole bunch of TRUEs.
lets convert the two year and month columns into a date
library(lubridate)
what will it look like if we
concatenate the year and month columns:
paste(utility$Year,
utility$Month, "01", sep="-")
lets create a new field using
that concatenation to create a date:
utility$newDate <-
ymd(paste(utility$Year, utility$Month, "01", sep="-"))
and then lets use ggplot2 to
take a look at some things like temperature, which should have a
familiar cyclical pattern and reasonable values for chicago
library(ggplot2)
ggplot(utility, aes(x=newDate, y=Temp_F)) + geom_point()
+ geom_line()
that should look pretty regular.
Now, depending on your screen, the plot may look a little small
which can make things hard to read but we can make the plot
wider and taller for all future graphs with a command like
options(repr.plot.width=20,
repr.plot.height=4)
and then I could take a look at
some of the utility data like natural gas to see if it follows
the same pattern while adding some better labels
ggplot(utility, aes(x=newDate, y=Gas_Th_per_Day)) +
geom_point() + geom_line() + labs(title = "Monthly Natural Gas
Usage", x = "Year", y ="Ave Therms per day")
hmmm ... here there is a general
pattern, and a time when that general pattern changes somewhat
I could draw both the
temperature and gas usage lines together and color the
temperature in red and the amount of gas used in blue. The gas
usage is also a much smaller number than the temperature so lets
scale it up by a factor of 10 before we display it. Since
natural gas is used for cooking and heating does the pattern
make sense?
ggplot(utility, aes(x=newDate, y=10*Gas_Th_per_Day)) +
geom_line(colour="blue") + geom_line(aes(y=Temp_F,
colour="red")) + xlab("Date") + labs(title = "Monthly Natural
Gas Usage vs Temperature", x = "Year", y ="Temp(F) & 10*Ave
Therms per day")
or take a look at electricity
and set the y axis lower limit to 0 and the upper limit to 60
ggplot(utility, aes(x=newDate, y=E_kWh_per_Day)) +
geom_line() + geom_point() + coord_cartesian(ylim =
c(0,60)) + labs(title = "Monthly Electricity Usage", x =
"Year", y ="Ave kWh per day")
since electricity is used for
air conditioning, among other things, there should be an overall
pattern that is similar to the natural gas usage, but like the
natural gas usage there are shifts in the overall patterns. Hmmm
... can electricity usage fall below zero? Yes it can.
we could just look at June
since that is a warm month
junes <- subset(utility, Month == 6)
ggplot(junes, aes(x=newDate, y=E_kWh_per_Day)) +
geom_point(color="blue") + geom_line(size=2) +
coord_cartesian(ylim = c(0,60)) + labs(title = "June
Electricity Usage", x = "Year", y ="Ave kWh per day")
maybe I want to compare June electricity usage to the
temperature in June over all these years to see if there is a
direct correlation
ggplot(data=junes, aes(x=newDate, y=E_kWh_per_Day,
colour="E_kWh_per_Day")) + geom_point() + geom_line(size=2) +
coord_cartesian(ylim = c(0,80)) + geom_line(aes(y=Temp_F,
colour="Temp_F"),size=2) + labs(title = "June Electricity
Usage Compared to Temperature", x = "Year", y ="kWh per day
& Temperature (F)")
For the rest of the class you should investigate the water, gas,
and electricity data to see what you can find, and create a
report in your groups Jupyter Notebook documenting your
findings. Remember doing this in science class? same thing, just
with data.
All of this from data on a
1-month time scale. As we move to this data being more available
on smaller timescales, down to 30 minutes for ComEd data, it
starts to become easier and easier to track people's behavior,
even down to knowing what room a person is likely in based on
real time utility usage (knowing when a gas oven turns on, or a
PC stars using more power). We will talk more about privacy
issues later in the course.
GitHub
If you don't have git on
your machine you can download it here - https://git-scm.com/
The
windows installer has a lot of options, but if you stick
to the defaults it should work fine. This will give you a
nice command line version and a primitive GUI version so
you may want to look into other GUI clients if that is
your style.
We will be using GitHub
for turning in the projects - https://github.com/
If you don't have a git
account yet you should sign up for one.
Some Tutorials
GitHub has their own
getting started page which is a nice intro - https://guides.github.com/activities/hello-world/
This is a nice intro for
the basics - https://rubygarage.org/blog/most-basic-git-commands-with-examples
Quick Command List - https://rogerdudler.github.io/git-guide/
At minimum you will be using
git to turn in all your code and data files from your
projects, but I would also recommend regularly updating your
files on git so there is some external proof of when you
submitted, as well as having backup copies at various
checkpoints. Note that git can also be a nice place to store a
copy of your website files to prove they were done on time,
and you can host your website for your project on git as well
if you prefer. Note that I would not rely solely on Git for
backing up your projects - keep multiple backups in multiple
places.
For example I could take my evl weather R project
above and move it up to a new git repository:
- I already have a Git account so I can use my
favorite web browser to go to github.com and sign in and use
the + in the upper right corner to add a New repository
- I can give it a name like Week2, make it private for now.
Then I click the green button to create the repository
- Keep this tab open in your browser - we will come back to
it.
- Go to the terminal and change the current
directory to the one for your R project, e.g. cd
Documents/CS424/evlWeatherForR
- If you look at the files in that directory you
should see a app.R, a jpg, some tsv files, etc. type git
init to create a new .git folder.
- Now we need to link this
local repository to GitHub so if you go back to GitHub and
look at the Code tab and then click on the green Code button
you can get the https link which will look something like https://github.com/YourGitAccountName/YourGitProjectName.git
- back in the terminal you can use the command git
remote add origin
https://github.com/YourGitAccountName/YourGitProjectName.git
so in my case git remote add origin https://github.com/andyevl/Week2.git
- You can then add all the files to the list to be
tracked with git add --all
- You can then commit the changes with an initial
comment (feel free to modify the comment) with git
commit -m 'starter project'
- You can check the status with git
status. Status is helpful to tell you which files
are being tracked, and which are untracked, which have been
modified, etc.
- Another helpful command with the
October 2020 switch from 'master' to 'main' as the
primary branch name is git branch.
This gives you the name of the branch you are linked to.
After October 2020 it should be 'main'. if it is
'master' then a handy command is git branch
-mv master main to rename the branch to
main in order to match Git's new default.
- Now we can finally push our code up onto GitHub
with git push origin main
- This will be
slightly unhappy since it wants a comment but you can just
save out of the editor.
- If you now go back to GitHub and refresh the
page you should see a lot more files there with their
modification dates.
- Whenever you want to update the repository at
GitHub with your latest version you would go to the top
level of your project directory and type this sequence to
update only those files that have changed (or added to the
repository):
- git
add --all
- git
commit -m 'new comment for this version'
- git
push origin main
Git has limits on file sizes (100 MB uploaded from
the command line and 25 MB uploaded from the browser) and
the maximum sizes of repositories (2 GB). The R code for the
projects will be small, but the data files will be getting
larger as the projects go on and you will need to make sure
that you can upload your data files on GitHub, which will
very likely mean trimming down the size of your data files,
breaking them into pieces, and possibly storing them in a
binary format, all of which will also help your dashboards
run faster. This is another reason to get a running version
of your code onto GitHub early so you don't need to deal
with breaking up your data file(s) near the project
deadline. GitLFS (large file storage) exists but has been
unreliable in past classes so it is NOT an option for this
course.
For the rest of this week I'd like people to get R and R studio
installed, and if you have not used R before then start with
Swirl
https://swirlstats.com/
If you are familiar with R then take a look at the shiny video
tutorials, which will be really helpful in getting ready for the
course projects, at
https://shiny.rstudio.com/tutorial/
By the end of this week you
should get the evl weather example above running in your local
copy of R Studio and then create a shinyapps.io account and
move the file up there so you can see it running on the web.
Add another page to your gradescope submission containing the
URL of your shinyapps.io site.

