BranchingSets: Interactively Visualizing Categories on Node-Link Diagrams
September 24th, 2016
Categories: Visual Analytics, Visual Informatics
Authors
Paduano, F., Etemadpour, R., Forbes, A.G.About
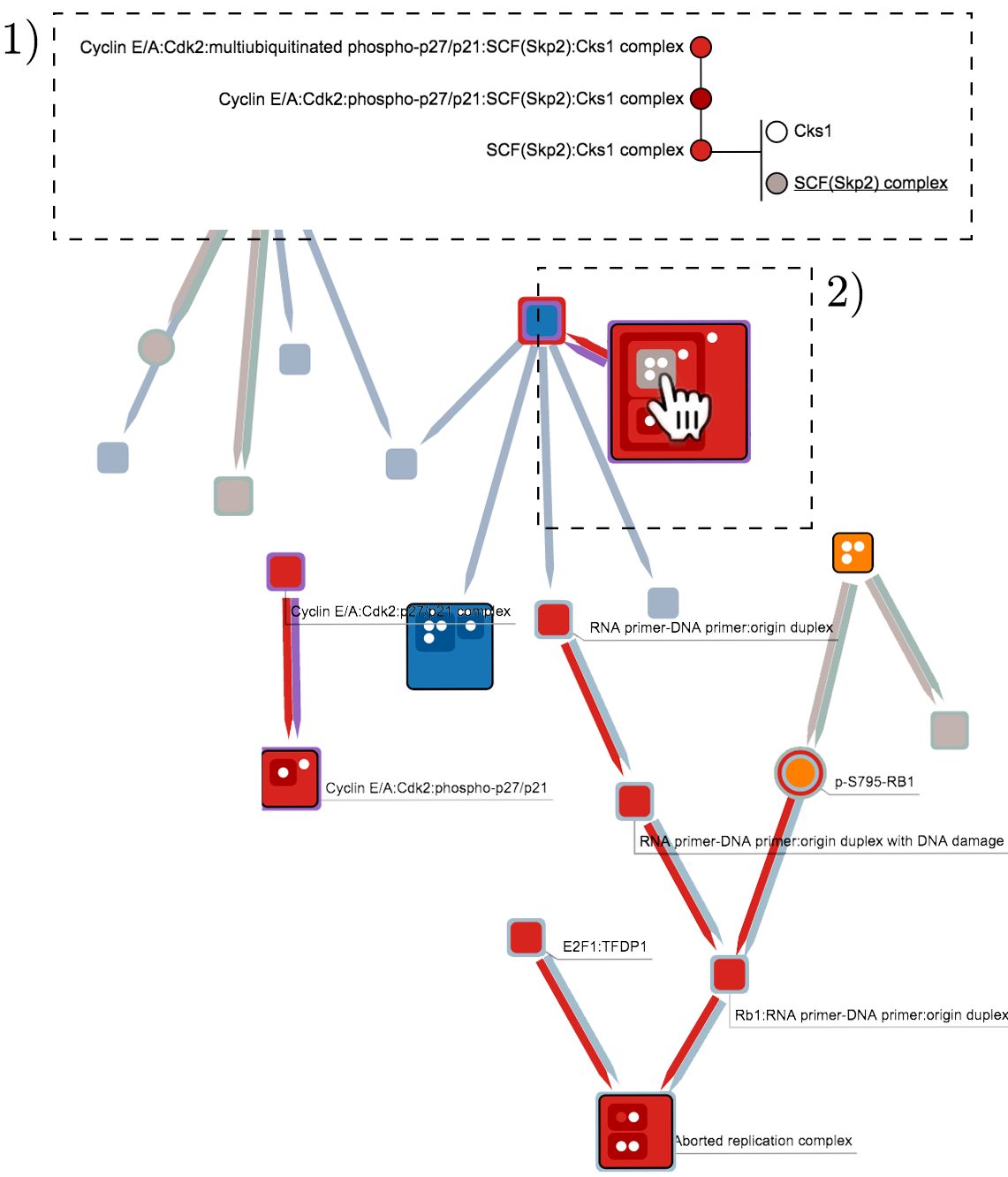
Node-link diagrams are widely used for visualizing relational data in a wide range of fields. However, in many situations it is useful to provide set membership information for elements in networks. We present BranchingSets, an interactive visualization technique that uses visual encodings similar to Kelp Diagrams in order to augment traditional node-link diagrams with information about the categories that both nodes and links belong to. BranchingSets introduces novel user-driven methods to procedurally navigate the graph topology and to interactively inspect complex, hierarchical data associated with individual nodes. Results indicate that users find the technique engaging and easy to use. This is further confirmed by a quantitative study that compares the effectiveness of the visual encodings used in BranchingSets to other techniques for displaying set membership within node-link diagrams, finding our technique more accurate and more efficient for facilitating interactive queries on networks containing nodes that belong to multiple sets.
Resources
URL
Citation
Paduano, F., Etemadpour, R., Forbes, A.G., BranchingSets: Interactively Visualizing Categories on Node-Link Diagrams, 9th International Symposium on Visual Information Communication and Interaction (VINCI 2016), Dallas, TX, September 24th, 2016. http://dx.doi.org/10.1145/2968220.2968229