Fixing TIM: Identifying Functional Mutations in Protein Families through the Interactive Exploration of Sequence and Structural Data
October 13th, 2013
Categories: Applications, Visual Analytics, Visual Informatics
Authors
Wenskovitch, J., Luciani, T., Chen, K., Marai. G. E.About
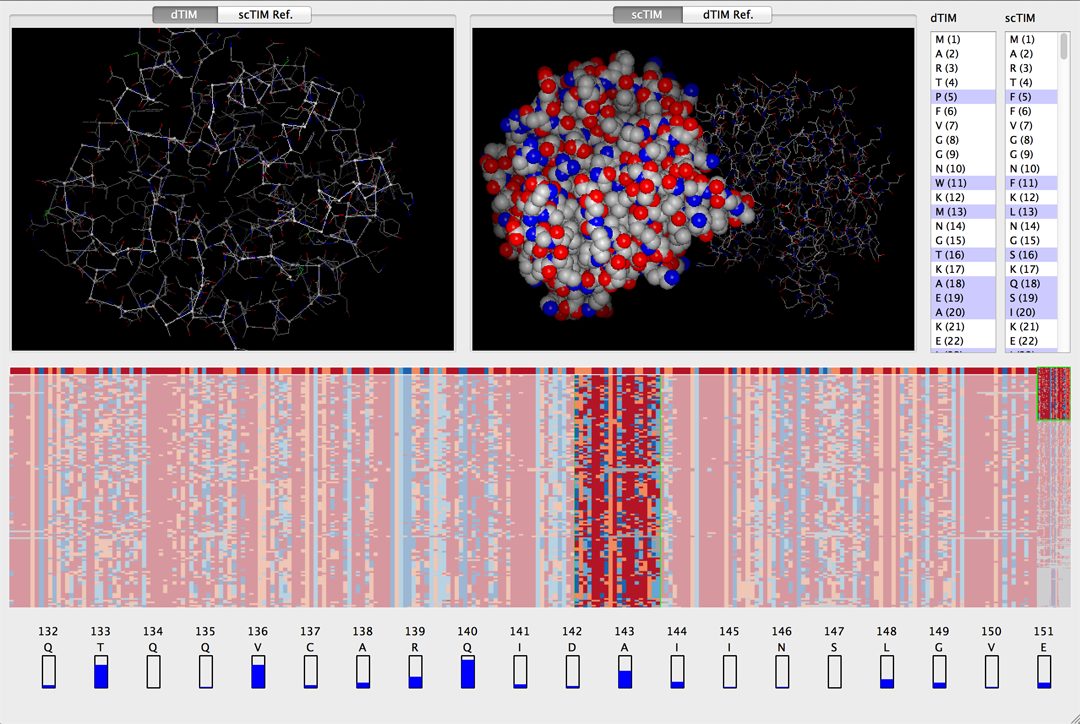
We present the design and implementation of a visual mining and analysis tool to help identify protein mutations across family structural models, and to help discover the effect of these mutations on protein function. We follow a client-server approach in which distributed data sources for 3D structure and non-spatial sequence information are seamlessly integrated into a common visual interface. Multiple linked views and a computational backbone allow comparison at the molecular and atomic levels, while a trend-image visual abstraction allows for the sorting and mining of large collections of sequences and of their residues. We evaluate our tool on the triosephosphate isomerase (TIM) family structural models and sequence data, and show that our tool provides an effective, scalable way to navigate a family of proteins, as well as a means to inspect the structure and sequence of individual proteins.
Keywords: Molecular Sequence Analysis, Molecular Structural Biology, Computational Proteomics
Resources
Citation
Wenskovitch, J., Luciani, T., Chen, K., Marai. G. E., Fixing TIM: Identifying Functional Mutations in Protein Families through the Interactive Exploration of Sequence and Structural Data, IEEE BioVis 2013 Data Competition, pp. 1-4, October 13th, 2013.