Evaluating the Effect of Right-Censored End Point Transformation for Radiomic Feature Selection of Data From Patients With Oropharyngeal Cancer
November 28th, 2018
Categories: Applications, Software, User Groups, Visual Analytics
Authors
Zdilar, L., Vock, D., Marai, G.E., Fuller, C.D., Mohamed, A.S.R., Elhalawani, H., Elgohari, B., Canahuate, G.About
Purpose
To evaluate the effect of transforming a right-censored outcome into binary, continuous, and censored-aware representations on radiomics feature selection and subsequent prediction of overall survival (OS) and relapse-free survival (RFS) of patients with oropharyngeal cancer.
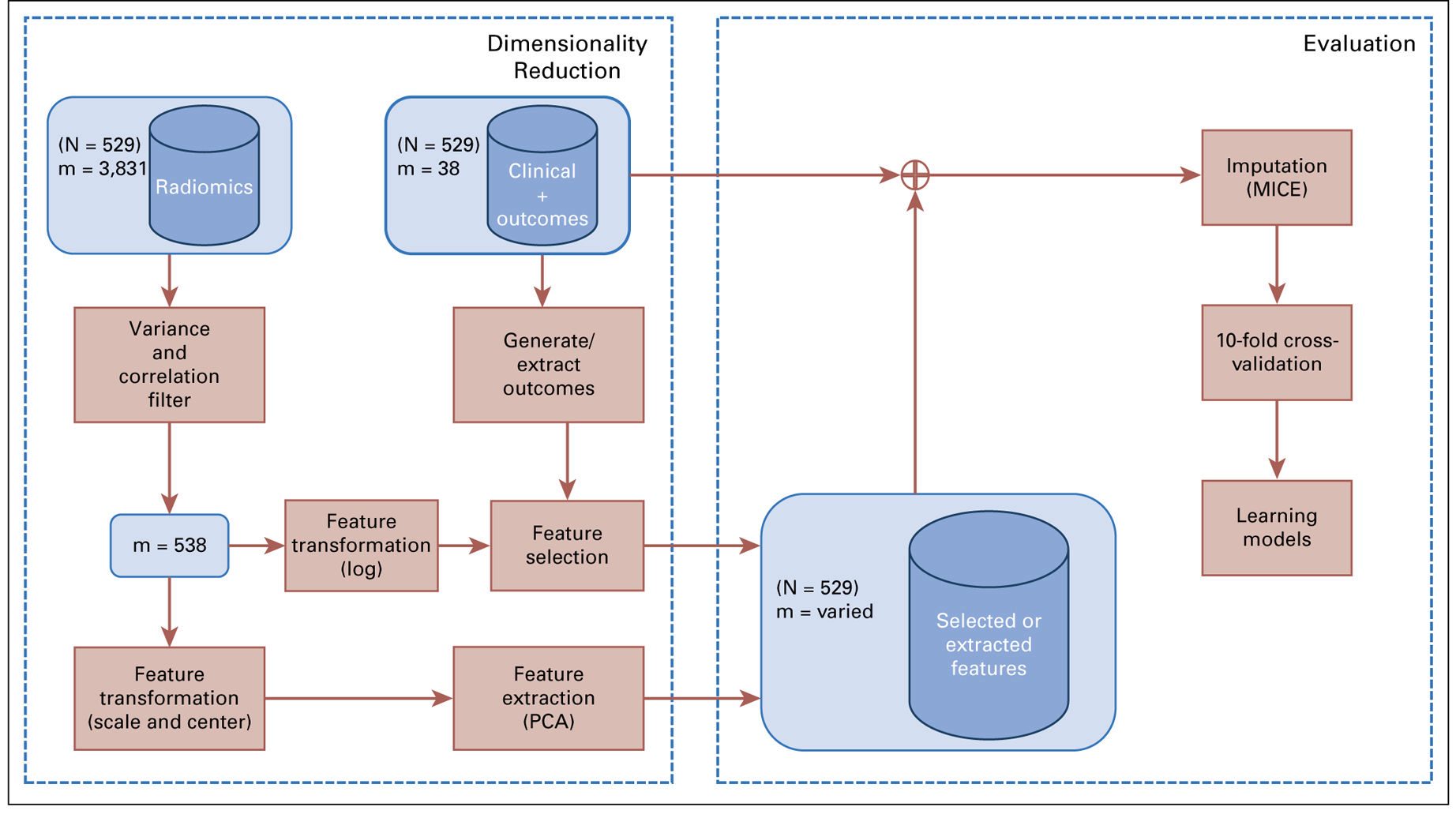
Methods and Materials
Different feature selection techniques were applied using a binary outcome indicating event occurrence before median follow-up time, a continuous outcome using the Martingale residuals from a proportional hazards model, and the raw right-censored time-to-event outcome. Radiomic signatures combined with clinical variables were used for risk prediction. Three metrics for accuracy and calibration were used to evaluate eight feature selectors and six predictive models.
Results
Feature selection across 529 patients on more than 3,800 radiomic features resulted in increases ranging from 0.01 to 0.11 in C-index and area under the curve (AUC) scores compared with clinical features alone. The ensemble model yielded the best scores for AUC and C-index (often > 0.7) and calibration (Nam-D’Agostino test statistic often < 15.5 with 8 df). The random forest feature selectors achieved the best performance considering all metrics. Random regression forest performed the best in OS prediction with the ensemble model (AUC, 0.75; C-index, 0.76; calibration, 8.7). Random survival forest performed the best in RFS prediction with the ensemble model (AUC, 0.71; C-index, 0.68; calibration, 19.1).
Conclusion
Including a radiomic signature results in better prediction than using only clinical data. Signatures generated randomly or without considering the outcome result in poor calibration scores. The random forest feature selectors for each of the three transformations typically selected the greatest number of features and produced the best predictions at acceptable calibration levels. In particular, random regression forest and random survival forest performed best for OS and RFS, respectively.
Resources
URL
Citation
Zdilar, L., Vock, D., Marai, G.E., Fuller, C.D., Mohamed, A.S.R., Elhalawani, H., Elgohari, B., Canahuate, G., Evaluating the Effect of Right-Censored End Point Transformation for Radiomic Feature Selection of Data From Patients With Oropharyngeal Cancer, JCO Clinical Cancer Informatics, no 2, pp. 1-19, November 28th, 2018. https://doi.org/10.1200/CCI.18.00052